

Overview
Every species on Earth has more than 20 different tRNAs. Each tRNA is made of a single RNA chain that base pairs with itself in a pattern consistent across all tRNAs. The tertiary structure of tRNA is characteristically viewed from an angle such that it resembles an inverted L, formed from two main base-paired helices which run perpendicular to each other.
Stacking interactions
Many of the bases in the helices are stacked, meaning that the aromatic rings of the bases are involved in hydrophobic interactions that act to stabilize the tertiary structure of tRNA.
AA acceptor stem and anticodon
Arguably the most fundamental landmarks of the tRNA structure are the attachment site for the amino acid it carries, and the anticodon bases. The amino acid acceptor region and the anticodon region are 76 Å apart. These regions are recognized by the enzymes that attach the amino acid corresponding to the anti-codon to the acceptor stem of different tRNAs.
All amino acid acceptor stems have the sequence "CCA", which occurs at the 3' end of the RNA chain. The amino acid (not shown) specified by the tRNA's anticodon will be attached to the 3'-OH group of the terminal adenine. The enzymes that attach the amino acids are called aminoacyl tRNA synthetases. There is a specific aminoacyl tRNA synthetase for each amino acid.
The 5' end of the tRNA chain is quite close to the 3' end in the tertiary structure.
tRNA secondary structure shows four base-paired stems or "arms" common to all tRNAs:
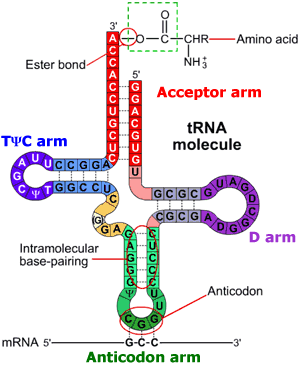
However, the secondary structure regions are not obvious in the inverted L-shape of a tRNA's tertiary structure. We'll "map" them onto the tertiary structure using color:
The anticodon arm
The anticodon arm contains the 3 base anticodon that pairs with a complementary codon in a messenger RNA. Recall that codons in mRNA specify particular amino acids. In this tRNA, the anticodon is A-A-mG (methyl-Guanine) which will pair with the codon U-U-C on an mRNA (not shown). UUC encodes the amino acid phenylalanine, which this tRNA will carry.
The D arm
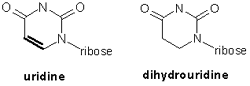
The D arm is named for an unusual nucleotide it contains, dihydrouracil, which is a modified uracil. In the drawing below, you can see that the difference between dihydrouridine and uridine (the base moiety of uracil) is the conversion of a double bond to a single bond. This means that two hydrogen atoms are added to the base, one to each atom at the end of the single bond. However, since structures solved using x-ray crystallography do not include the position of the hydrogen atoms, we can show you where the modified base is in this 3D model of tRNA, but the modification itself will not be visible.
The variable arm
The variable arm has between 3 and 21 nucleotides, depending on which amino acid the tRNA encodes. This tRNA's variable arm is very short so it looks quite different from the other arms of the molecule. In some cases, the length of the variable arm is important in the recognition of the aminoacyl tRNA synthetase for the tRNA.
The TψC arm
The TψC arm contains the nucleotide pseudouracil, which is symbolized by the greek letter ψ (psi, pronounced like the word "sigh"). Look closely at the diagram below. Can you verify that the highlighted nucleotide in the animation that follows is pseudouracil as opposed to uracil?
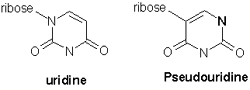
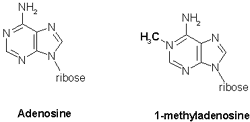
1-methyl-adenine is a modified adenine nuceotide found in tRNA:
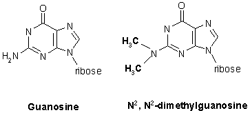
N2,N2-dimethylguanine is a modified guanine:
Non-standard base-pairing
In addition there are several non-Watson-Crick base pairs in tRNA that take part in cross-linking in the nonhelical regions of tRNA. These are tertiary interactions that involve unusual pairs such as A-A, A-C, and G-G, for example. An A-G pair is shown below. Dashed lines indicate hydrogen bonds.
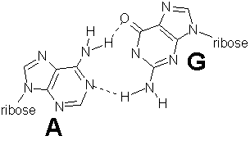
Some of these base pairs help to stabilize the interactions between different arms. For example, a G on the D arm pairs with a ψ on the TψC arm.
For specific instructions on how to manipulate the 3D images in this tutorial, see Structure Tutorial Help.