 Introduction
Introduction
DNA contains all the information for encoding the amino acids that make up proteins. DNA is made up of nucleotides, and proteins are made up of amino acids. The information in DNA is first transcribed into messenger RNA (mRNA), which is also made up of nucleotides. The process of converting the genetic message found in mRNA to an amino acid sequence (protein synthesis) is called translation. How does the cell convert the nucleotide sequence of mRNA into the amino acid sequence of proteins? This process involves a large enzyme called the ribosome and an adapter molecule between the two languages of mRNA sequence and peptide sequence called transfer RNA (tRNA).
The Ribosome
The synthesis of proteins is catalyzed by the ribosome. The ribosome is made up of a large and small subunit, and is a large enzyme comprised mostly of ribosomal RNA (rRNA), with proteins interspersed like islands in a sea of RNA. Besides the rRNA, the ribosome contains binding sites for tRNA and mRNA. The rRNA forms most of the ribosomal structure and performs the catalytic steps of peptide synthesis, the mRNA delivers the genetic message, and tRNA translates the genetic code into peptide sequence.
Transfer RNA
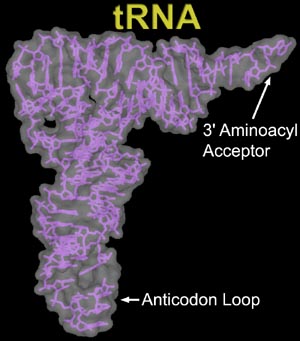
tRNA is involved in the translation of the nucleic acid message into the amino acids of proteins. tRNA itself is an RNA molecule with a conserved inverted L structure. One end of the tRNA contains an anticodon loop which pairs with a mRNA specifying a certain amino acid. The other end of the tRNA has the amino acid attached to the 3' OH group via an ester linkage.
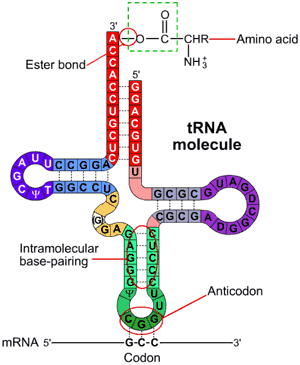 tRNA with an attached amino acid is said to be "charged". The enzyme that attaches the amino acid to the 3'-OH is called an aminoacyl tRNA synthetase (aaRS). There is a specific tRNA for each amino acid, 20 in all. Similarly, there is a specific aaRS for each tRNA.
tRNA with an attached amino acid is said to be "charged". The enzyme that attaches the amino acid to the 3'-OH is called an aminoacyl tRNA synthetase (aaRS). There is a specific tRNA for each amino acid, 20 in all. Similarly, there is a specific aaRS for each tRNA.
Only the first 2 nucleotides in the tRNA anticodon loop are strictly required for the decoding of the mRNA codon into an amino acid. The third nucleotide in the anticodon is less stringent in its base-pairing to the codon, and is referred to as the "wobble" base. Since the genetic code is degenerate, meaning that more than one codon can specify a single amino acid, the anticodon of tRNA can pair with more than one mRNA codon and still be specific for a single amino acid.
Protein synthesis begins when a charged tRNA, mRNA, and the small and large ribosomal subunits (30S and 50S for E. coli) come together to form the initiation c complex (70 S in E. coli). The initiation complex contains a peptidyl binding site, (P site) and an aminoacyl binding site (A site). The first tRNA to bind is always the initiator tRNA, tRNAfMET. tRNAfMET binds to the start codon of mRNA, AUG. The first amino acid of the protein is thus methionine. Elongation of the polypeptide chain begins when the anticodon of a second charged tRNA binds to the next mRNA codon in the vacant A site of the 70S initiation complex. The second charged tRNA is guided to the A site by elongation factors. The P and the A sites are very close together. This allows the carboxy terminus of the amino acid in the P site to react with the nucleophilic amino terminus of the amino acid on the tRNA in the A site to form a stable peptide bond. This reaction is catalyzed by peptidyl transferase, a ribozyme associated with the 50S subunit. The tRNA in the P site is now devoid of its amino acid and the tRNA in the A site is now a dipeptidyl tRNA with 2 amino acids. The 70S complex now moves along the mRNA toward its 3' end. This movement is called translocation and causes the tRNA in the P site to be displaced. The dipeptidyl tRNA in the A site moves into the P site so that another charged tRNA can move into the A site. Each translocation step is coupled to the hydrolysis of GTP. Elongation continues in this fashion until the stop codon is reached and the peptide chain is released from the ribosome.
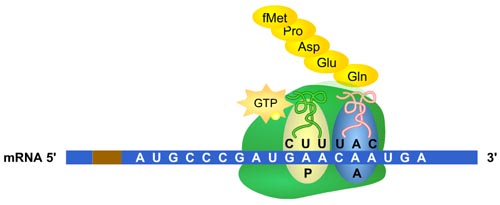 |
Translation of proteins involves a mRNA, tRNA, ribosome, and energy.
Click on the graphic to learn about translation with the Protein Synthesis animation.
|
Important points to remember are:
-
tRNAs have an inverted L structure
-
tRNAs have an anticodon loop that pairs with a mRNA codon
-
there is a tRNA for each amino acid
-
amino acids are attached to tRNAs via ester linkage to the 3'-OH by specific aaRS to form charged tRNAs
-
charged tRNAs pair with the mRNA codons in the P and A sites of the 70S initiation complex
-
peptide chain elongation proceeds when the attached amino acid of the tRNA in the P site forms a peptide bond with the amino acid of the tRNA in the A site
The crystal structure of tRNA elucidates other important points:
-
tRNAs all have an inverted L structure
-
tRNAs have conserved structural features: 5'P, D arm, anticodon arm, variable arm, TyC arm, 3' amino acid acceptor stem
-
tRNAs contain unusual bases
-
tRNAs are made up of a single chain of RNA yet have considerable tertiary structure arising from base pairing within the chain
-
there are several non-Watson-Crick base pairs that take part in cross-linking
-
tRNAs are narrow so as to be able to sit next to one another on adjacent mRNA codons in the P and A sites
-
the anticodon arm and amino acid acceptor stems are solvent accessible regions of the molecule




 Introduction
Introduction tRNA with an attached amino acid is said to be "charged". The enzyme that attaches the amino acid to the 3'-OH is called an aminoacyl tRNA synthetase (aaRS). There is a specific tRNA for each amino acid, 20 in all. Similarly, there is a specific aaRS for each tRNA.
tRNA with an attached amino acid is said to be "charged". The enzyme that attaches the amino acid to the 3'-OH is called an aminoacyl tRNA synthetase (aaRS). There is a specific tRNA for each amino acid, 20 in all. Similarly, there is a specific aaRS for each tRNA.